This is the first issue of my journal of theoretical neuroscience.
Editorial
This month, I have selected 4 papers on spike initiation (1-4), 1 classical paper on the theory of brain energetics (5), and 1 paper on bibliometrics (6). Three of the papers on spike initiation (1-3) have in common that they are about the relation between geometry (morphology of the neuron and spatial distribution of channels) and excitability. Spikes are initiated in a small region called the axon initial segment (AIS), and this region is very close to the soma. Thus there is a discontinuity in both the geometry (big soma, thin axon) and the spatial distribution of channels (lots in the AIS). It has great impact on excitability, but this has not been very deeply explored theoretically. In fact, as I have discussed in a recent review (Brette, 2015), most theory on excitability (dynamical systems theory) has been developed on isopotential models, and so is largely obsolete. So there is much to do on spike initiation theory that takes into account the soma-AIS system.
Articles
1. Evans MD, Tufo C, Dumitrescu AS and MS Grubb. (2017). Myosin II activity is required for structural plasticity at the axon initial segment. (Comment on PubPeer)
A number of studies have shown that the AIS can move over hours or days, with various manipulations such as depolarizing the neuron (as in this study) or stimulating it optogenetically. Two open questions: what are the molecular mechanisms involved in this displacement? Is it actually a displacement or is it just that stuff is removed at one end and inserted at the other end? The same lab previously addressed the first question, showing the involvement of somatic L-type calcium channels and calmodulin. This study shows that myosin (the stuff of muscle, except not the type expressed in muscles) is involved, which strongly suggests that it is an actual displacement; this is in line with previous studies showing that dendrites and axons are contractile structures (e.g. Roland et al. (2014)). This and previous studies start to provide building blocks for a model of activity-dependent structural plasticity of the AIS (working on it!).
2. Hamada M, Goethals S, de Vries S, Brette R, Kole M (2016). Covariation of axon initial segment location and dendritic tree normalizes the somatic action potential. (Comment on PubPeer)
Full disclosure: I am an author of this paper. In the lab, we are currently interested in the relation between neural geometry and excitability. In particular, what is the electrical impact of the location of the axon initial segment (AIS)? Experimentally, this is a difficult question because manipulations of AIS geometry (distance, length) also induce changes in Nav channel and other channel properties, in particular phosphorylation (Evans et al., 2015). So this is typically a good question for theorists. I have previously shown that moving the AIS away from the soma should make the neuron more excitable (lower spike threshold), everything else being equal (Brette, 2013). Here we look at what happens after axonal spike initiation, when the current enters the soma (I try to avoid the term “backpropagate”, see Telenczuk et al., 2016). The basic insight is simple: when the axonal spike is fully developed, the voltage gradient between soma and start of the AIS should be roughly 100 mV, and so the axonal current into the soma should be roughly 100 mV divided by resistance between soma and AIS, which is proportional to AIS distance. Next, to charge a big somatodendritic compartment, you need a bigger current. So we predict that big neurons should have a more proximal AIS. This is what the data obtained by Kole’s lab show in this paper (along with many other things, as our theoretical work is a small part of the paper – as often, most of the theory ends up in the supplementary).
3. Michalikova M, Remme MWH and R Kempter. (2017). Spikelets in Pyramidal Neurons: Action Potentials Initiated in the Axon Initial Segment That Do Not Activate the Soma. (Comment on PubPeer)
Using simulations of detailed models, the authors propose to explain the observation of spikelets in vivo (small all-or-none events) by the failed propagation of axonal spikes to the soma. Under certain circumstances, they show that a spike generated at the distal axonal initiation site may fail to reach the somatic threshold for AP generation, so that only the smaller axonal spike is observed at the soma. This paper provides a nice overview of the topic and I found the study convincing. There is in fact a direct relation to our paper discussed above (Hamada et al., 2016): this study shows how the axonal spike can fail to trigger the somatic spike, which explains why the AIS needs to be placed at the right position to prevent this. One can argue (speculatively) that if AIS position is indeed tuned to produce the right amount of somatic depolarization, then sometimes this should fail and result in a spikelet (algorithm: if no spikelet, move AIS distally; if spikelet, move AIS proximally).
4. Mensi S, Hagens P, Gerstner W and C Pozzorini (2016). Enhanced Sensitivity to Rapid Input Fluctuations by Nonlinear Threshold Dynamics in Neocortical Pyramidal Neurons. (Comment on PubPeer)
I had to love this paper, because the authors basically experimentally confirm every theoretical prediction we had made in our paper on spike threshold adaptation (Platkiewicz and Brette, 2011). Essentially, what we had done is derive the dynamics of spike threshold from the dynamics of Nav channel inactivation. There were a number of non-trivial predictions, such as the shortening of the effective integration time constant, sensitivity to input variance, the specific way in which spike threshold depends on membrane potential, and the interaction between spike-triggered and subthreshold adaptation (that we touched upon in the discussion). This study uses a non-parametric model-fitting approach in cortical slices to empirically derive the dynamics of spike threshold (indirectly, based on responses to fluctuating currents), and the results are completely in line with our theoretical predictions.
5. Attwell D and SB Laughlin (2001). An energy budget for signaling in the grey matter of the brain (Comment on PubPeer and Pubmed Commons).
This is an old but important paper on energetics of the brain, in particular: how much does it cost to maintain the resting potential? How much does it cost to propagate a spike? The paper explains some theoretical ideas to do these estimations, and is also a good source for relevant empirical numbers. It is important though to look at follow-up studies, which have addressed some issues, for example action potential efficiency is underestimated in this study. One problem in this study is the estimation of the cost of the resting potential, which I think is just wrong (see my detailed comment on Pubmed Commons; I believe the authors will soon respond). Unfortunately, I think it is really hard to estimate this cost by theoretical means; it would require knowing the permeability at rest to various ions, most importantly in the axon.
6. Brembs B, Button K and M Munafò (2013). Deep Impact: Unintended consequences of journal rank. (Comment on PubPeer)
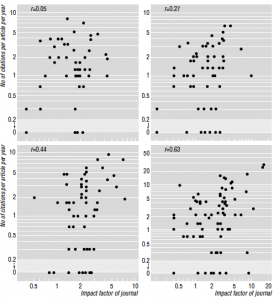
The authors look at the relation between journal rank (derived from impact factor) and various indicators, for example effect sizes reported, statistical power, etc. In summary, they found that the only thing journal rank strongly correlates with is the proportion of retractions and frauds. Another interesting finding is about the predictive power of journal rank on future citations. There is obviously a positive correlation since impact factor measures the number of citations. But it is really quite small (see my post on this). What is most interesting is that the predictive power started increasing in the 1960s, when the impact factor was introduced. This strongly suggests that, rather than being a quality indicator, the impact factor biases the citations of papers (increases the visibility of otherwise equally good papers). This paper also shows evidence of manipulation of impact factors by journals (including Current Biology, whose impact factor went from 7 to 12 after its acquisition by Elsevier), and is generally a good source of references on the subject.